In the war on antibiotic-resistant bacteria, it's not so much the antibiotics that are making the enemy stronger as it is how they are prescribed. A new study suggests that doctors can beat antibiotic resistance using those same antibiotics but in a very targeted manner and in combination with other health strategies.
The current broad application of antibiotics helps resistant bacterial strains evolve forward. But using data about bacteria’s specific resistances when prescribing those same drugs more precisely can help put the evolution of resistant strains in reverse, according to researchers from the Georgia Institute of Technology, Duke University, and Harvard University who conducted the study.
One researcher cautioned that time is pressing: New strategies against resistance that leverage antibiotics need to be in place before bacteria resistant to most every known antibiotic become too widespread. That would render antibiotics nearly useless, and it has been widely reported that this could happen by mid-century, making bacterial infections much more lethal.
“Once you get to that pan-resistant state, it’s over,” said Sam Brown, who co-led the study and is an associate professor in Georgia Tech’s School of Biological Sciences. “Timing is, unfortunately, an issue in tackling antibiotic resistance.”
The new study, which was co-led by game theorist David McAdams, a professor of business administration and economics at Duke University, delivers a mathematical model to help clinical and public health researchers devise new concrete prescription strategies and those supporting health strategies. The measures center on the analysis of bacterial strains to determine what drugs they are resistant to, and which not.
Some medical labs already scan human genomes for hereditary predispositions to certain medical conditions. Bacterial genomes are far simpler and much easier to analyze, and though the analytical technology is currently not standard equipment in doctors’ offices or medical labs they routinely work with, the researchers think this could change in a reasonable amount of time. This would enable the study’s approach.
The researchers published their study in the journal PLOS Biology on May 16, 2019. The work was funded by the Centers for Disease Control and Prevention, the National Institute of General Medical Sciences, the Simons Foundation, the Human Frontier Science Program, the Wenner-Gren Foundations, and the Royal Physiographic Society of Lund.
Q&A
Here are some questions and answers on how the study’s counterintuitive approach could beat back antibiotic resistance:
Isn’t prescribing antibiotics the problem? How can it fight resistance?
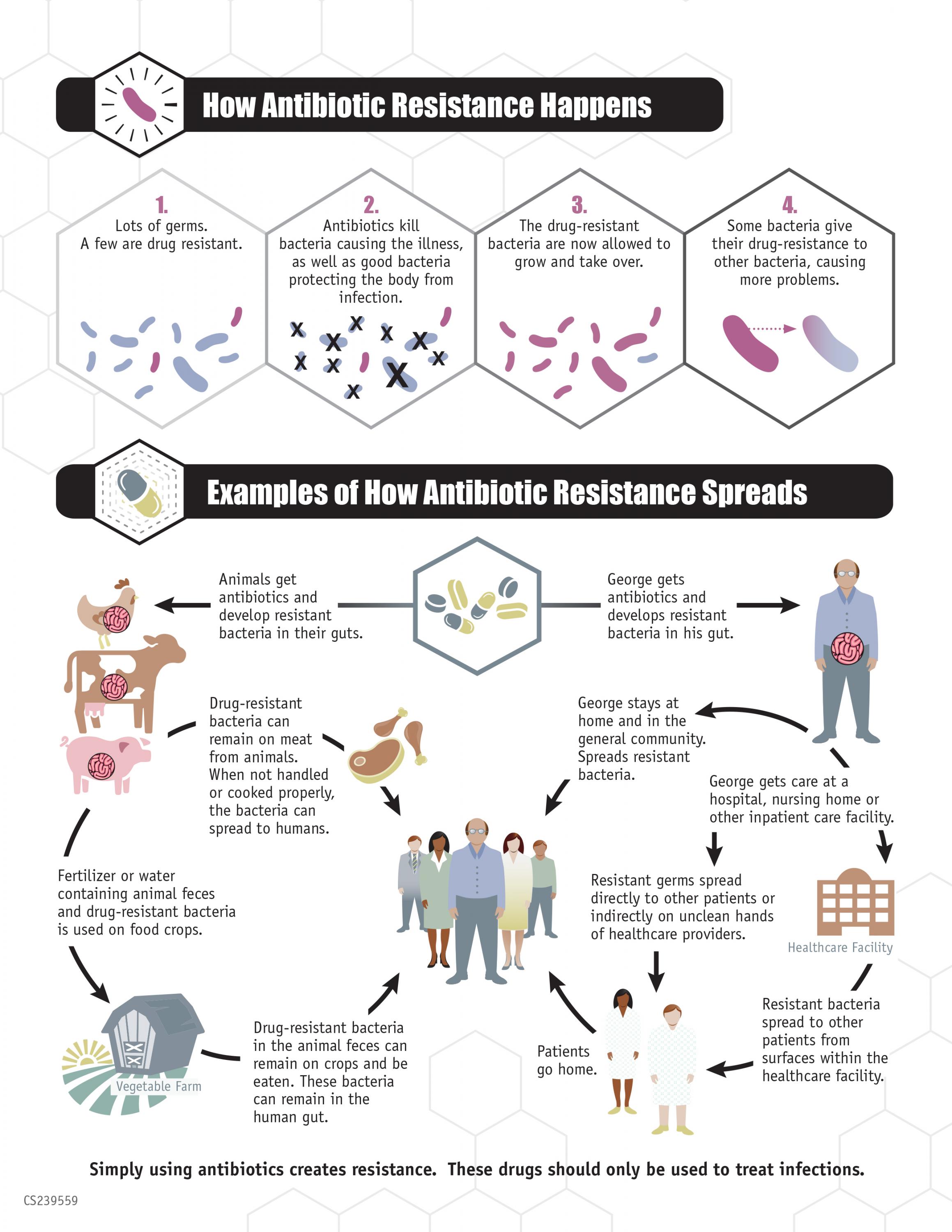
The real problem is the broad application of antibiotics. They treat human infections and farm animals, and in the process are killing off a lot of non-resistant bacteria while bacteria resistant to those drugs survive. The resistant strains can then reproduce and with fewer competitors in their space, then they dominate bacterial communities in the host animals and people.
The resistant bacteria get passed to other hosts and become more prevalent in the world altogether. New prescription strategies would outsmart that evolutionary scenario by exposing through genomic (or other) analysis bacteria’s resistance but also their vulnerabilities.
“Right now, there are rapid tests for the pathogen. If you’ve got strep throat, the clinic swabs the bacteria and does a rapid assay that says yes, that’s streptococcus,” Brown said. “But it won’t tell you if it’s resistant to the drug usually prescribed against it. In the future, diagnostics at the point-of-care could find out what strain you’ve got and if it’s resistant.”
Then clinicians would choose the specific antibiotics that the bacteria are not resistant to, and kill the bacteria, thus also stopping them from spreading the genes behind their resistance to other antibiotics. So, identifying an infector’s resistance hits two birds with one stone.
“It’s great for fighting antibiotic resistance, but it’s also good for patients because we’ll always use the correct antibiotic,” Brown said.
[Thinking about grad school? Here's how to apply to Georgia Tech.]
Are there enough effective antibiotics left to do this with?
Plenty. Antibiotics still work as a rule.
In addition, searching out and destroying resistant bacteria could help refresh existing antibiotics’ effectiveness.
“The idea is prevalent that we will use antibiotics up, and then they’re gone,” Brown said. “It doesn’t have to be that way. This study introduces the concept that antibiotics could become a renewable resource if we act on time.”
As mentioned above, prescription strategies by themselves won’t beat resistance, right?
Correct. Resistance evolution has some tricky complexities.
“A lot of bacteria with the potential to make us sick like E. coli spend most of their time just lurking peacefully in our bodies. These are bystander bacteria, and they are exposed to lots of antibiotics that we take for other things such as sore throats or ear aches,” Brown said. “This frequent exposure is probably the major driver of resistance evolution.”
The antibiotic prescription strategy needs those additional health care measures to win the fight, but those measures are pretty straightforward.
What are those additional measures?
Diagnostics need to apply to bystander bacteria, too. E. coli in the intestine or, for example, Strep pneumoniae living peacefully in nostrils would be checked for resistance, say, during annual checkups.
“If the patient is carrying a resistant strain, you work to beat it back before it can break out,” Brown said. “There could be non-antibiotic treatments that do this like, perhaps, bacteria replacement.”
Bacteria replacement therapy would introduce new bacteria into the patient’s body to outcompete the undesirable antibiotic-resistant bacteria and displace it. Also, people would stay home from school and work for a few days so as not to spread the bad bacteria to other people while their immune systems and possibly alternative therapies, such as bacteriophages or non-antibiotic drugs battle the bad bacteria.
This sounds hopeful, but are there other real-world circumstances to consider?
“The study’s mathematical models are broad simplifications of real life,” Brown said. “They don’t take into account that pathogens spend a lot of time in other antibiotic-exposed environments such as farms. Dealing with that is going to require some more research.”
The study also purposely leaves out "polymicrobial infections," which are infections by multiple kinds of bacteria at the same time. The researchers believe that the study’s models can still be relevant to them.
“We expect the logic of combating drug resistance to still hold in these more complex scenarios, but diagnostics and treatment rules will have to be honed for them specifically,” Brown said.
Also read: Want to beat antibiotic resistance? Rethink that strep throat prescription
These researchers coauthored the study: David McAdams from Duke University, Kristofer Wollein Waldetoft from Georgia Tech, and Christine Tedijanto and Marc Lipsitch from Harvard University. The research was funded by the Centers for Disease Control and Prevention (grant OADS BAA 2016-N-17812), the National Institute of General Medical Sciences at the National Institutes of Health (grant U54GM088558), the Simons Foundation (grant 396001), the Human Frontier Science Program (grant RGP0011/2014), the Wenner-Gren Foundations, and the Royal Physiographic Society of Lund.
Media contact/writer: Ben Brumfield
(404) 660-1408
Research News
Georgia Institute of Technology
177 North Avenue
Atlanta, Georgia 30332-0181 USA