From Parkinson’s and Alzheimer's to cardiac arrhythmia, amyloids are linked to a number of diseases. These aggregates of proteins form in the body when a protein loses its normal structure and misfolds or mutates. And since many of these proteins are large and complicated, just how some of these mutations occur and aggregate remains a mystery — as does the creation of effective treatments.
New research on glaucoma led by Georgia Tech chemists and an alumna may help change that.
“There has been a lot of work done to understand how smaller folded proteins form amyloid aggregates, but this study helps us to understand the aggregation pathway of a larger, more complex system,” says co-first author Emily Saccuzzo. That work could one day help scientists uncover new modes of treatment — not just for glaucoma, but for other diseases caused by protein aggregation, as well.
Saccuzzo started the project in 2018 as a graduate student in the Lieberman Lab in the School of Chemistry and Biochemistry at Georgia Tech, and is now a Postdoctoral Research Associate at Pacific Northwest National Labs. “Emily was a summer student before she matriculated, and she established the initial feasibility of doing these experiments,” says Raquel Lieberman, professor and Sepcic Pfeil Chair in Chemistry at Georgia Tech. “I'm immensely proud of her.”
Their research team's recent findings are featured in a new paper, “Competition between inside-out unfolding and pathogenic aggregation in an amyloid-forming β-propeller," published in the journal Nature Communications.
Lieberman and Saccuzzo brought together researchers from throughout and beyond the Institute to collaborate on the study.
“This was a very multi-disciplinary project, and that's always really satisfying,” Lieberman says. “I think when you bring more people to the table, you can answer hard questions and do more than you can do on your own.”
The Georgia Tech research team includes Hailee F. Scelsi, Minh Thu Ma, and Shannon E. Hill of the School of Chemistry and Biochemistry; Xinya Su and Matthew P. Torres of the School of Biological Sciences; Elisa Rheaume or the Interdisciplinary Graduate Program in Quantitative Biosciences; and James C. Gumbart, who holds joint appointments in the School of Chemistry and Biochemistry, School of Biological Sciences, and School of Physics. The research team also includes Saccuzzo's co-first author Mubark D. Mebrat, Minjoo Kim, and Wade D. Van Horn of Arizona State University as well as Renhao Li of the Emory University School of Medicine.
A complicated protein
While many studies have focused on smaller proteins, called model proteins, that have established ‘rules’ and known patterns for amyloid-formation (a specialized type of protein aggregation), the protein that contributes to glaucoma is larger and more complex. This type of larger, complicated protein is relatively unstudied.
“We had known for a while that mutations in myocilin can cause the protein to misfold and aggregate, which in turn leads to glaucoma,” Saccuzzo says. “What we didn’t know, however, was the exact mechanism by which this protein misfolds and aggregates.
“The goal of this study was to determine how disease mutants are misfolded, in hopes that that would give us insight into the early steps in the aggregation pathway,” she adds.
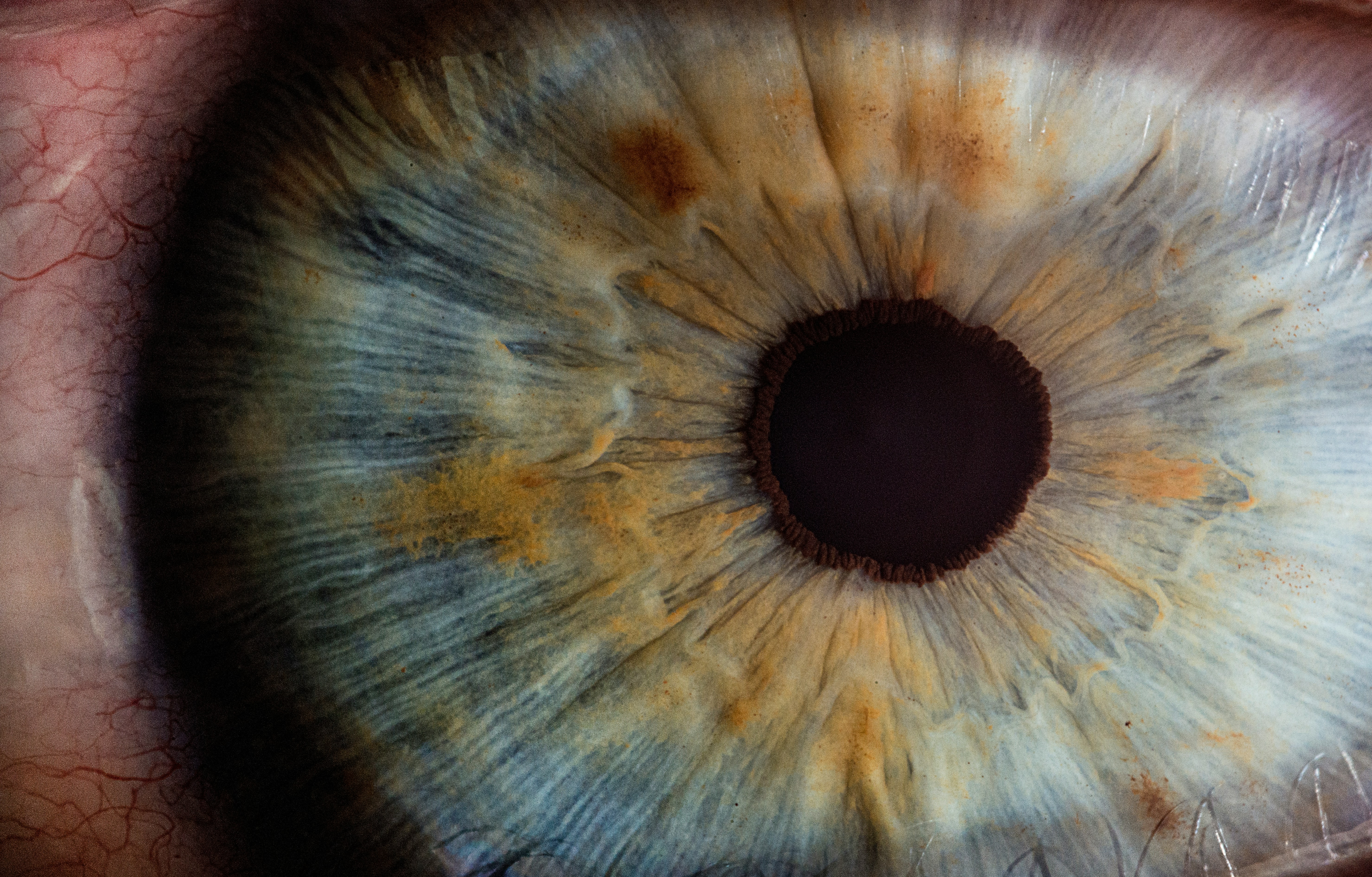
Located at the interface between the white of the eye and the colored iris, the protein forms a tiny small ring all the way around the eye. “Every time you blink, you stretch that muscle. Every time the wind blows really strong, or you get something in your eye. Every time you rub your eye, you could be affecting this protein — even when it's not causing disease,” Lieberman says. Still, scientists aren’t sure what the protein does. “We only know what it's doing when it's causing trouble,” like glaucoma, she explains. “We don't know what its actual biological function is.”
Lieberman was initially attracted to the idea of studying the protein because she wondered if the research done on the model proteins might be applicable to the protein causing glaucoma. “The really early studies showed that it was likely similar to these model proteins that form amyloid,” Lieberman says. “I wanted to look into that because if we could show that that was true, then we could tap into the amazing resources and research done on model systems to help us combat the disease.”
An unpredictable system
“This was one of the largest amyloid-forming proteins characterized to date,” Saccuzzo says, and while the team hoped that they would find similarities to model proteins, the larger glaucoma-associated protein showed increased complexity.
“I think one of the most surprising observations that we made is that the protein itself is not at equilibrium for about 90 days after it’s made,” Lieberman adds. “One of the tenets of protein chemistry is that amino acid sequences adopt a unique structure, and that all of the information needed to fold the protein into its 3D structure is held in that amino acid sequence.”
Here, the protein was shimmying a small amount, meaning that it wasn’t at equilibrium. “There's so much more going on in the system than anyone could have imagined,” Lieberman explains. “We assume that the shape controls some of the properties, but this is another mystery of this protein.”
Because the protein is so complicated and isn’t at equilibrium, “there is a long list of the things we can’t predict,” says Lieberman, adding that it makes computer predictions difficult, along with certain experiments. “That was a moment when we thought: wow, here's this new system that people should think about. The rules might be refined to help us better understand what's going on.”
The future of protein modeling
While further research will need to be conducted in order to determine how best to treat glaucoma, the study provides a critical foundation for future studies. “What is not clear to me right now is whether we would be able to find one drug for all the people who have mutations, or if we need a specific drug for each type of mutation that we would encounter,” Lieberman says. While the research doesn’t prove that one treatment might not be effective for all, “it certainly shows that there's a lot more to this system than we ever expected.”
“Understanding what disease mutants look like at the molecular level could help pave the way for structurally-specific glaucoma therapeutics and diagnostic tools,” Saccuzzo adds.
Lieberman and Saccuzzo also underscore that the work done to understand the protein responsible for glaucoma can also be applied to other large proteins.
“At the end of the day, more proteins are not model proteins than are model proteins,” Lieberman says. “There are many more systems out there, and I suspect that there are many more proteins that can aggregate and may contribute to disease or aging that have yet to be explored. I think this research shows the value of bringing lots of different approaches to probing a complicated system to learn more about it.”
DOI: https://doi.org/10.1038/s41467-023-44479-2
Research reported in this publication was supported by the National Institutes of Health award numbers R01EY021205 (RLL, WVH), R41EY031203 (RLL), R01GM123169 (JCG), and R35GM141933 (WVH). EGS, HFS, and MTM were supported in part by 5T32EY007092-35.
Raquel Lieberman's research is supported by the Kelly Sepcic Pfeil, Ph.D. Faculty Endowment Fund.


Written by Selena LangnerContact: Jess Hunt-Ralston
